CycPeptMPDB ID: 2360
Peptide Information
| Source | 2018_Naylor | ||||||||
| Original Name in Source Literature | CSA-iso(67) | ||||||||
| Permeability 1 | -7.40 (PAMPA) | ||||||||
| Detection Limit of Permeability 1 | N.D | ||||||||
| Permeability 2 | -6.78 (RRCK) | ||||||||
| Detection Limit of Permeability 2 | N.D | ||||||||
| Molecular Weight | 1202.63 | ||||||||
| Monomer Length | 11 | ||||||||
| Molecule Shape | Circle | ||||||||
| EPSA | N.D | ||||||||
| Other Sources |
|
Structural Information
| Structure |
WARNING
3D structure on the left is the minimum energy conformation
obtained by the force field of molecular mechanics.
This conformation likely does not reflect what is found in biological systems,
and most peptides populate ensembles rather than a single conformation.
|
| Canonical SMILES | C/C=C/C[C@@H](C)[C@H]1OC(=O)[C@H](C(C)C)N(C)C(=O)[C@H](CC(C)C)N(C)C(=O)[C@H](CC(C)C)N(C)C(=O)[C@@H](C)NC(=O)[C@H](C)NC(=O)[C@H](CC(C)C)N(C)C(=O)[C@H](C(C)C)NC(=O)[C@H](CC(C)C)N(C)C(=O)CN(C)C(=O)[C@H](CC)NC(=O)[C@H]1NC |
| Sequence (HELM) |
PEPTIDE2360
{[Me_Bmt(E)].[Abu].[Sar].[meL].V.[meL].A.[dA].[meL].[meL].[meV]} $PEPTIDE2360,PEPTIDE2360,1:R3-11:R2$$$ 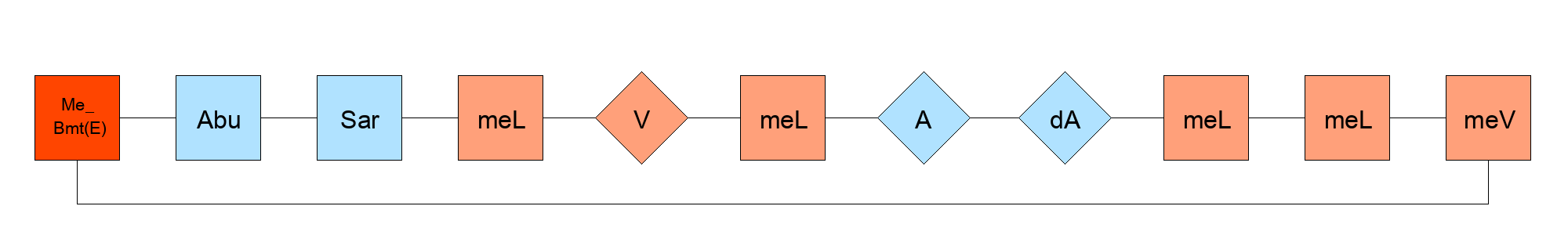
|
Physicochemical Properties
| LogP | 3.58 | (Calculated by RDKit) |
| Ring Count | 1 | (Calculated by RDKit) |
| Heavy Atom Count | 85 | (Calculated by RDKit) |
| Hydrogen Bond Acceptor Count | 13 | (Calculated by RDKit) |
| Hydrogen Bond Donor Count | 5 | (Calculated by RDKit) |
| Topological Polar Surface Area | 276.59 | (Calculated by RDKit) |
| 3D Polar Surface Area in Chloroform | 207.00 | (Calculated by Dr. Richard A. Lewis) |
| 3D Polar Surface Area in Water | 208.00 | (Calculated by Dr. Richard A. Lewis) |