Usage
Selector
Please Select a Section of Usage
1. Home Page
1.1 Introduction
Cyclic peptides have lately been expected as a new medical modality because
they can target "undruggable" targets such as intracellular protein-protein interactions.
However, cyclic peptides tend to have
poor membrane permeability due to their large size,
which is the biggest obstacle to successful drug discovery.
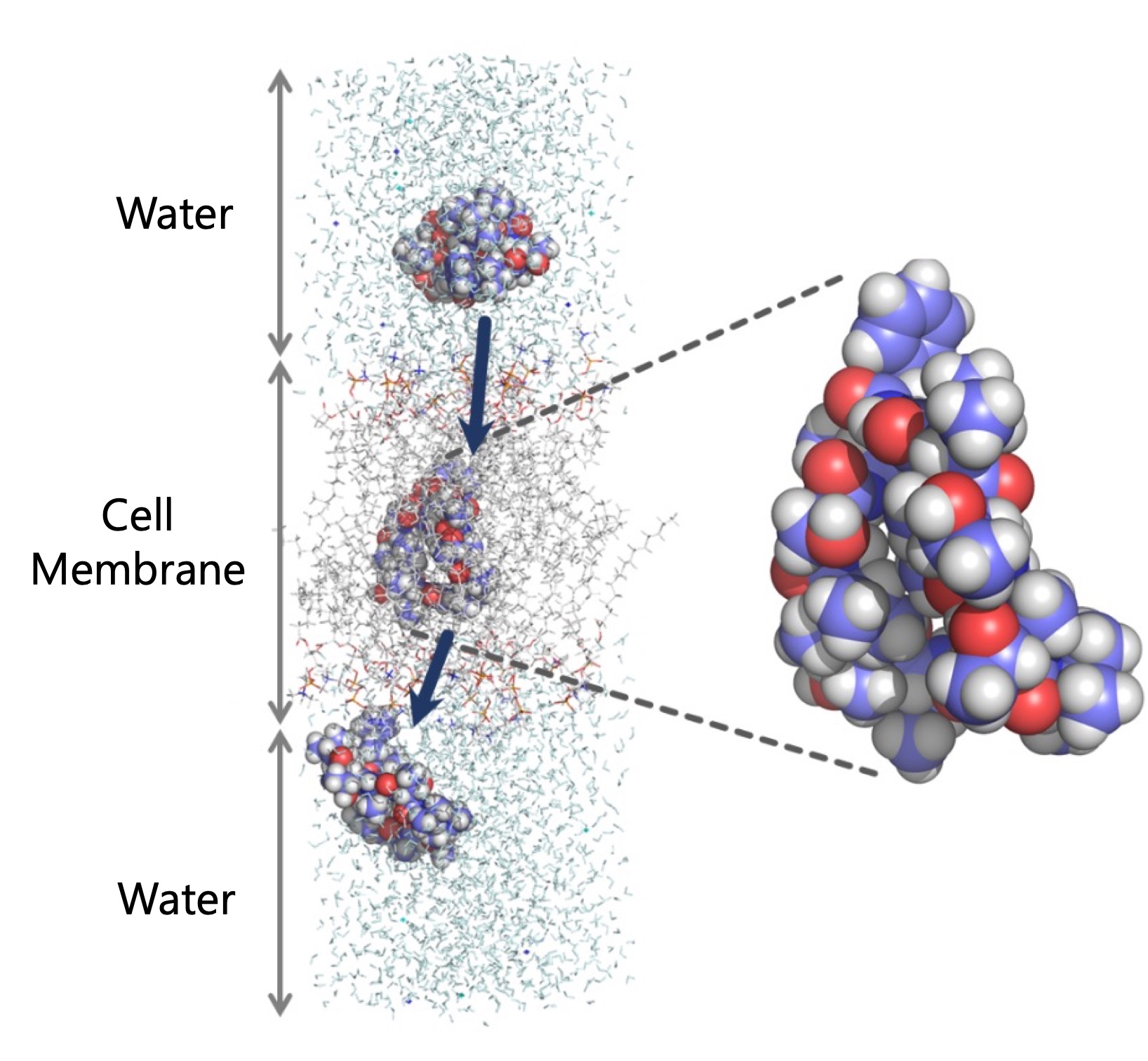
CycPeptMPDB (Cyclic Peptide Membrane Permeability Database) is a
comprehensive database of membrane permeability for cyclic peptides.
We not only collected the structure and membrane permeability information of cyclic peptides from many publications,
but also unified sequence notations that can express cyclic peptides concisely using HELM (Hierarchical Editing Language for Macromolecules) notation.
CycPeptMPDB is mainly composed of two parts: cyclic peptides and their constituent monomers (substructures).
1.2 Search Panel
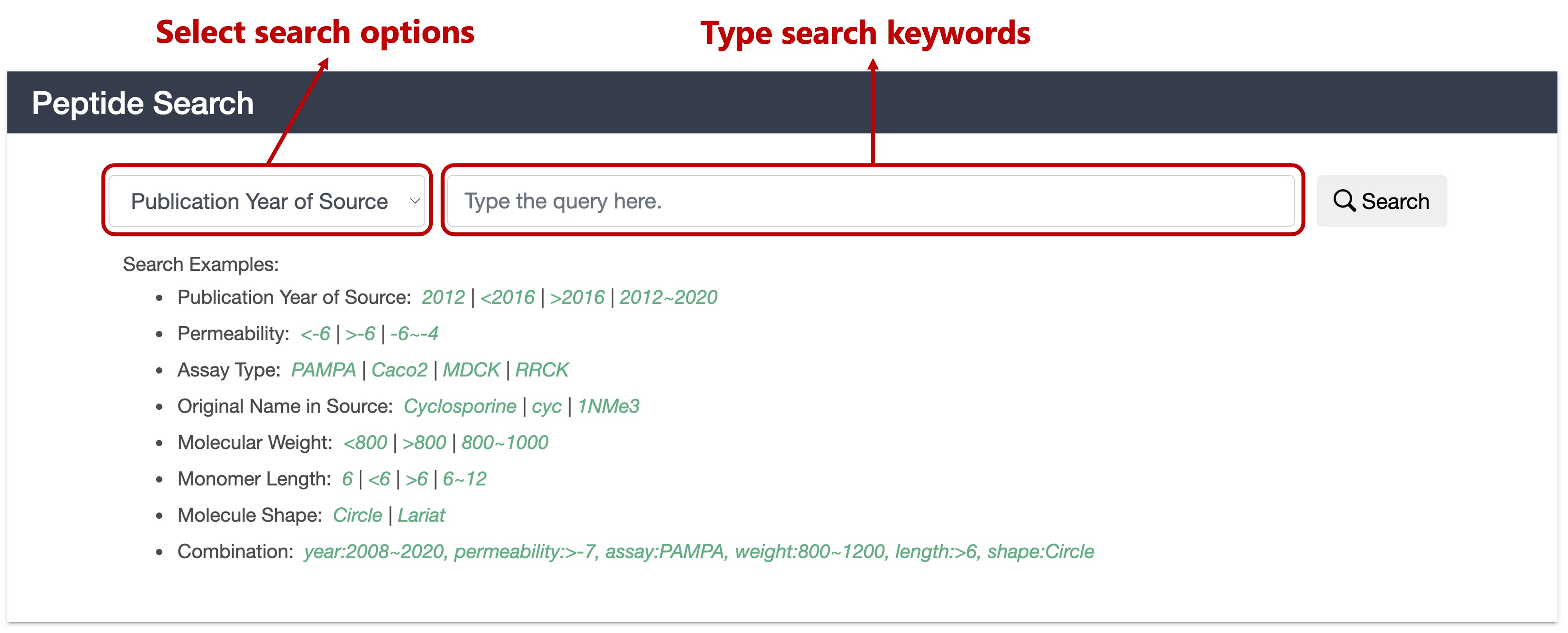
- Publication Year of Source: Search by the publication year of source. You can search for specific year (2012) or ranges (<2016, >2016, 2012~2020).
- Permeability: Search by the log scaled experimental permeability of the peptide. You can search for specific permeability (-6.4) or ranges (<-6, >-6, -6~-4).
- Assay Type: Search by the permeability assays type. There are four types of search candidates: PAMPA (parallel artificial membrane permeability assay), Caco2 (Caco-2 cell permeability assay), MDCK (Madin-Darby canine kidney cell permeability assay) and RRCK (Ralph Russ canine kidney cell, or called Low Efflux Madin-Darby canine kidney cell permeability assay).
- Original Name in Source: Search by compound name (Cyclosporine, 1NMe3) or part of name (cyc) of a peptide described in the source publication.
- Molecular Weight: Search by the molecular weight of the peptide (MolWt descriptor calculated by RDKit software). You can search for specific molecular weight (800) or ranges (<800, >800, 800~1000).
- Monomer Length: Search by the number of monomers that make up the peptide (sequence length). You can search for specific length (6) or ranges (<6, >6, 6~12).
- Molecule Shape: Search by the molecule shape of the peptide. There are two types of search candidates: Circle (cyclization position is the N- and C-terminal of sequence) and Lariat (cyclization position is not at the terminal of the sequence, namely one or more monomers in the side chain).
- Combination: Search by any combination of the above seven options. For queries, write the name of each option (year:, permeability:, assay:, name:, weight:, length:, shape:) and separate them with commas.

2. Peptide Browse
2.1 Browsing
CycPeptMPDB currently contains 7,991 structurally diverse cyclic peptides
collected from 56 publications. Some peptides overlapped in structure between
different publications and had different membrane permeability measurements,
they were recorded as separate data in CycPeptMPDB
(there were 8,466 peptides including duplicated structures).
In addition, all measured values were recorded when there were measurements
by multiple assays in one publication.
Therefore, the sum of the number of data in each subset when classified by Assay
Type is greater than the total number.
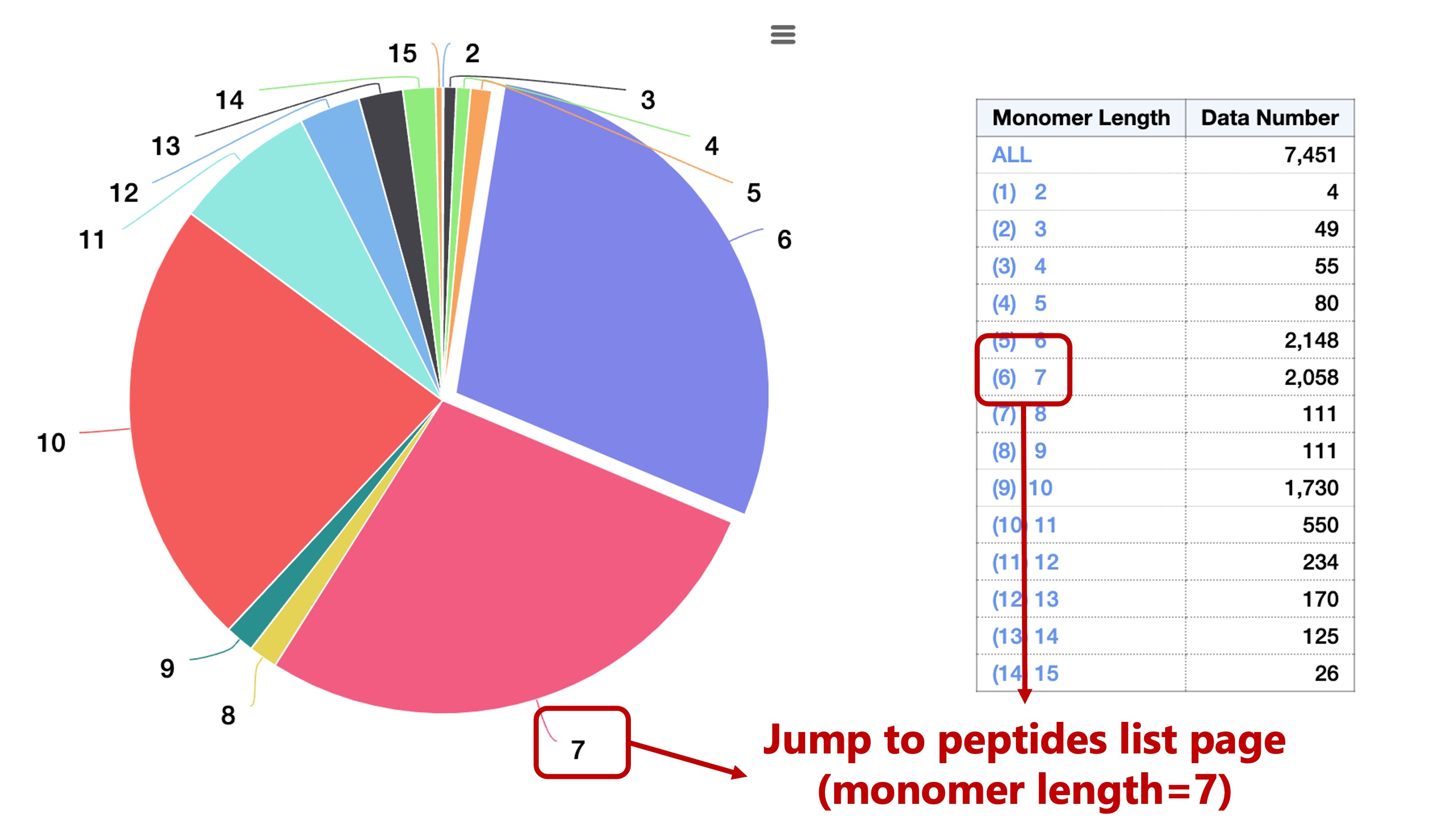
These peptides were classified in four ways:
 You can then select the corresponding subset from the pie chart labels or from the table on the right:
You can then select the corresponding subset from the pie chart labels or from the table on the right:


2.2 Peptide List
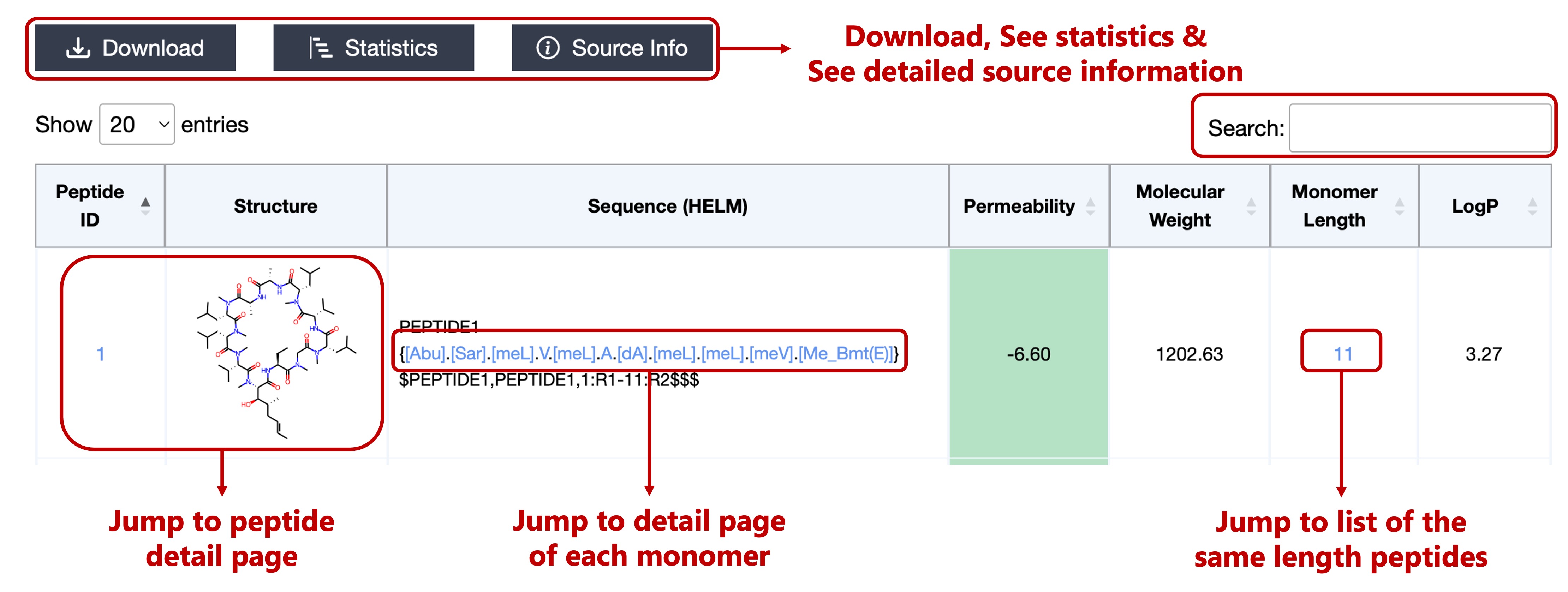
- CycPeptMPDB ID
- 2D Structure Image
- HELM Notation
- Permeability
- Molecular Weight (MolWt descriptor calculated by RDKit software)
- Monomer Length
- LogP (MolLogP descriptor calculated by RDKit software)
If permeability < -6.00, the background color of the cell is green.
If -6.00 ≤ permeability, the background color of the cell is yellow.
If you want to narrow down the peptides further, you can use the Search function
on the upper right of the table. However, the Search function here
differs from the Search Panel described in Section 1.2 in that
it can filter peptides that partially match the contents of the table
(in addition to the contents shown, data source name, publication year of source,
original name in source, and molecule shape are also search target).
Click on the Peptide ID or 2D Structure Image can jump to the peptide detail page.
Click on each monomer in the Sequence can jump to the detail page for that monomer.
Click on the Monomer Length can jump to a list of the same length peptides.
2.3 Peptide Detail
This page contains three sections: Peptide Information,
Structural Information and Physicochemical Properties.
2.3.1 Peptide Information:
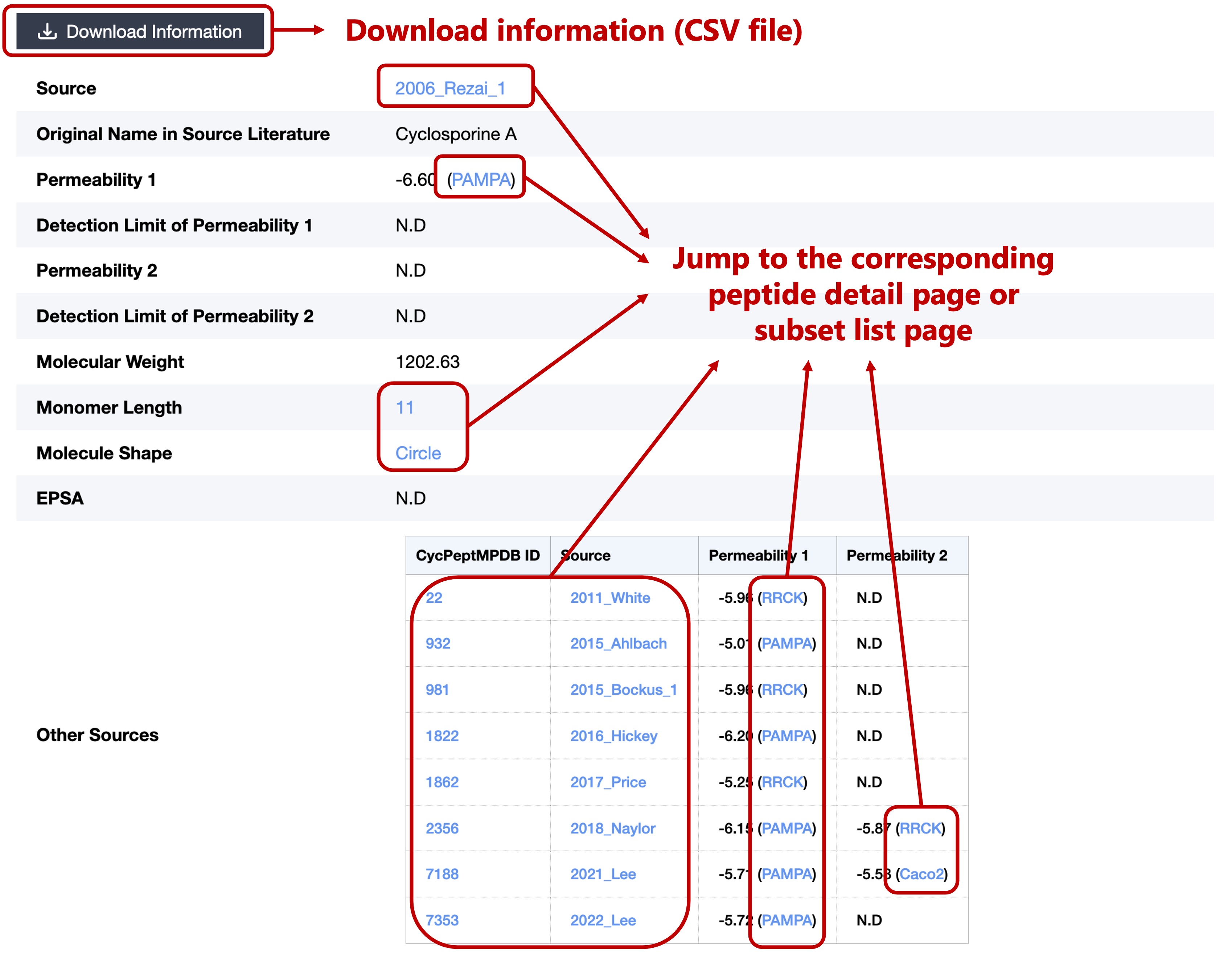 This section contains the basic information of each peptide, including:
This section contains the basic information of each peptide, including:

- Source
- Original Name in Source Literature
- Permeability (if the source mentioned %R and %T they will also be displayed)
- Detection Limit (if there was a description in the source)
- Molecular Weight
- Monomer Length
- Molecule Shape
- EPSA (if there was a measured value in the source)
- Other Sources (peptides with the same structure)
2.3.2 Structural Information:
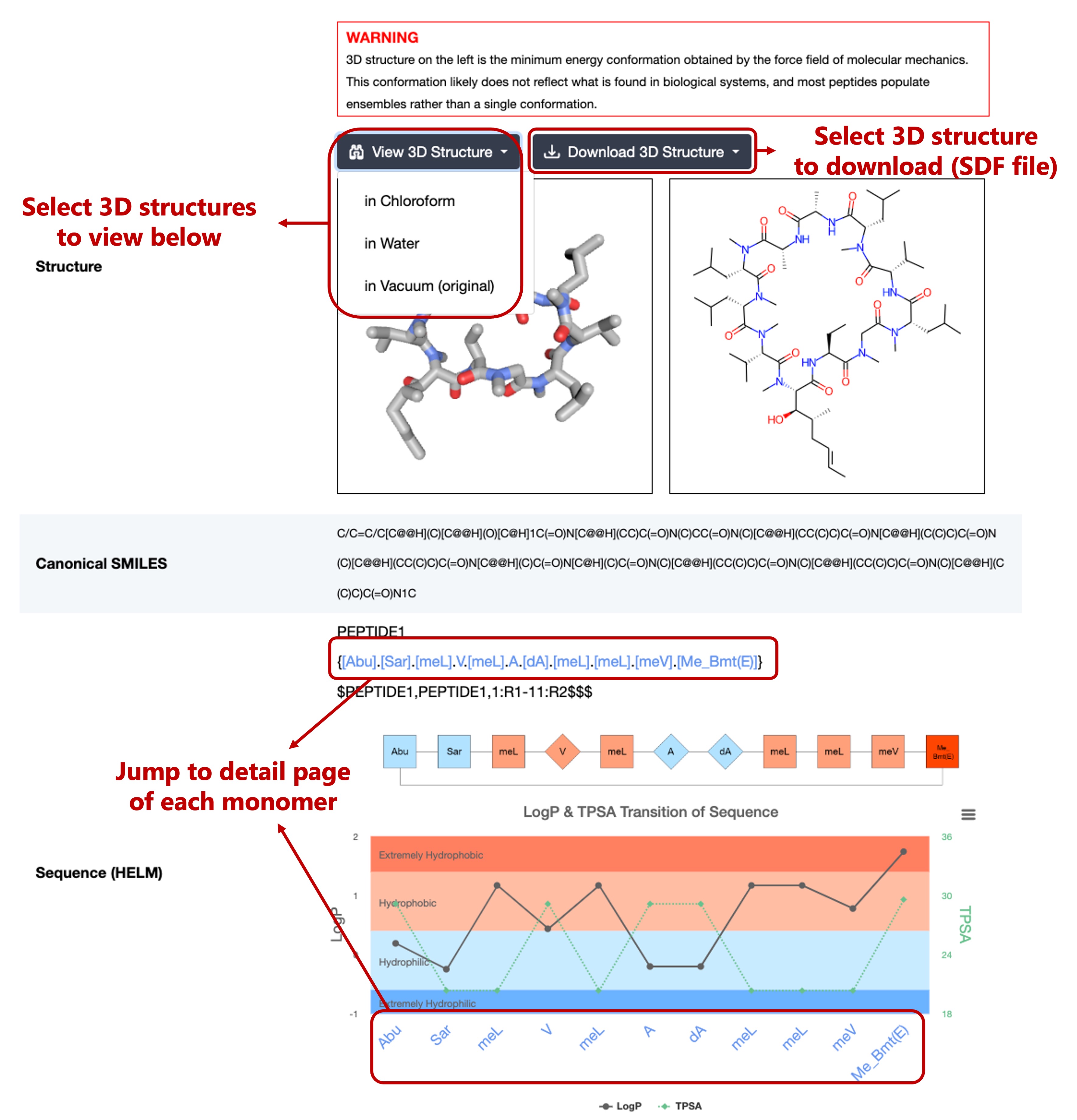 This section contains the structural information of each peptide, including:
This section contains the structural information of each peptide, including:

- Structure (3D structure view and 2D structure image)
- (version 1.1) Added 3D structures of cyclic peptides in chloroform and in water selected based on the score given by a fast QM method.
- Canonical SMILES
- Sequence (HELM)
2.3.3 Physicochemical Properties:
This section contains 8 types of descriptor for each peptide, including:
This section contains 8 types of descriptor for each peptide, including:
- LogP (MolLogP descriptor, calculated by RDKit software)
- Ring Count (RingCount descriptor, calculated by RDKit software)
- Heavy Atom Count (HeavyAtomCount descriptor, calculated by RDKit software)
- Hydrogen Bond Acceptor Count (NumHAcceptors descriptor, calculated by RDKit software)
- Hydrogen Bond Donor Count (NumHDonors descriptor, calculated by RDKit software)
- Topological Polar Surface Area (TPSA descriptor, calculated by RDKit software)
- (version 1.1) 3D Polar Surface Area in Chloroform (calculated by Dr. Richard A. Lewis)
- (version 1.1) 3D Polar Surface Area in Water (calculated by Dr. Richard A. Lewis)
3. Monomer Browse
3.1 Browsing
We defined the partial structure obtained after cleaving the peptide bonds and ester bonds
of the cyclic peptide as a monomer
(CycPeptMPDB has no data containing disulfide bonds).
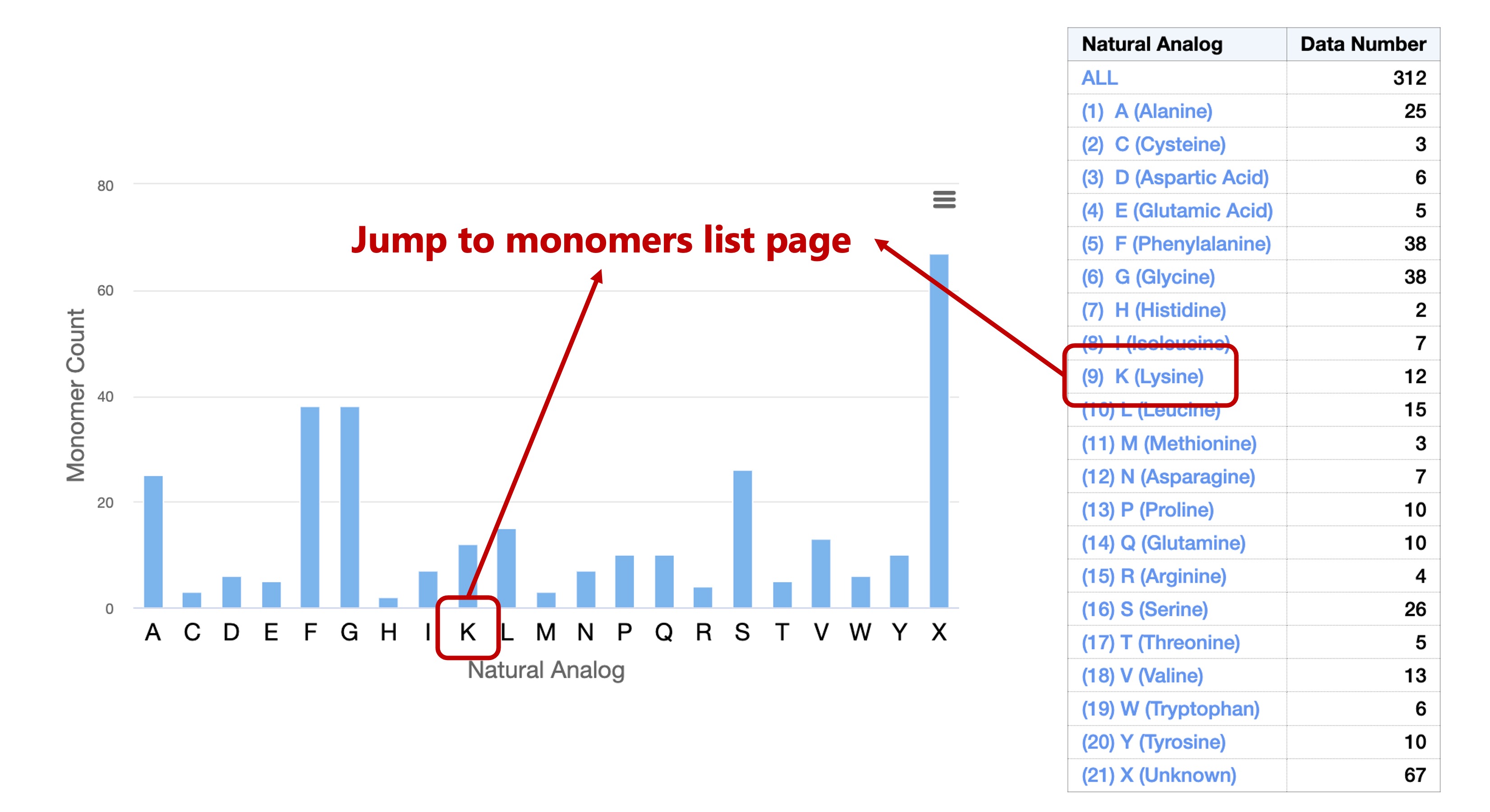
As a result, a total of 385 types of monomers were obtained.
These monomers were divided into 21 types according to their Natural Analog
(20 types of natural amino acids + unknown).
This classification referenced the description of each monomer in PubChem and the monomer library of ChEMBL.
You can then select the corresponding subset from the chart labels or from the table on the right:

3.2 Monomer List
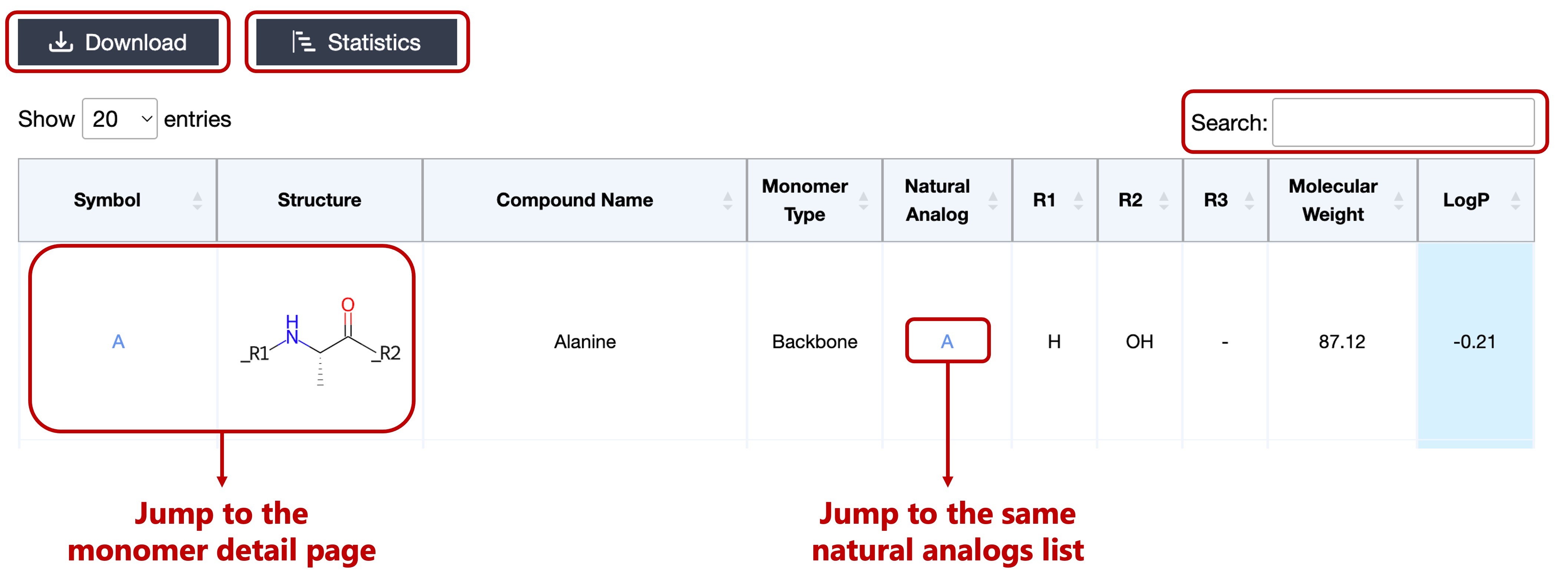
- Symbol (notation name in HELM)
- 2D Structure Image
- Monomer Type (Backbone or Terminal)
- Natural Analog
- Attachment Points (R1~R3)
- Molecular Weight (MolWt descriptor calculated by RDKit software)
- LogP (MolLogP descriptor calculated by RDKit software)
On the other hand, when calculating descriptors for monomers (e.g., alanine),
the presence of hydrogen bond donors and acceptors for the amide group and carboxyl group
may not accurately represent the partial physicochemical properties of the cyclic peptide before cleavage.
Therefore, if the original attachment point atom was H, H was replaced with methyl (CH3),
and if the original attachment point atom was OH, OH was deleted.
All monomer descriptors shown in CycPeptMPDB were calculated from such processed molecules.
If LogP < -0.60, the background color of the cell is blue.
If -0.60 ≤ LogP < 0.40, the background color of the cell is light blue.
If 0.40 ≤ LogP < 1.40, the background color of the cell is orange.
If 1.40 ≤ LogP, the background color of the cell is red.
If -0.60 ≤ LogP < 0.40, the background color of the cell is light blue.
If 0.40 ≤ LogP < 1.40, the background color of the cell is orange.
If 1.40 ≤ LogP, the background color of the cell is red.
If you want to narrow down the monomers further, you can use the Search function
on the upper right of the table.
It can filter monomers that partially match the contents of the table.
Click on the Symbol or 2D Structure Image can jump to the monomer detail page.
Click on the Natural Analog can jump to a list of the same analogs.
3.3 Monomer Detail
This page contains three sections:
Monomer Information,
Peptides Containing Current Monomer
and Physicochemical Properties.
3.3.1 Monomer Information:
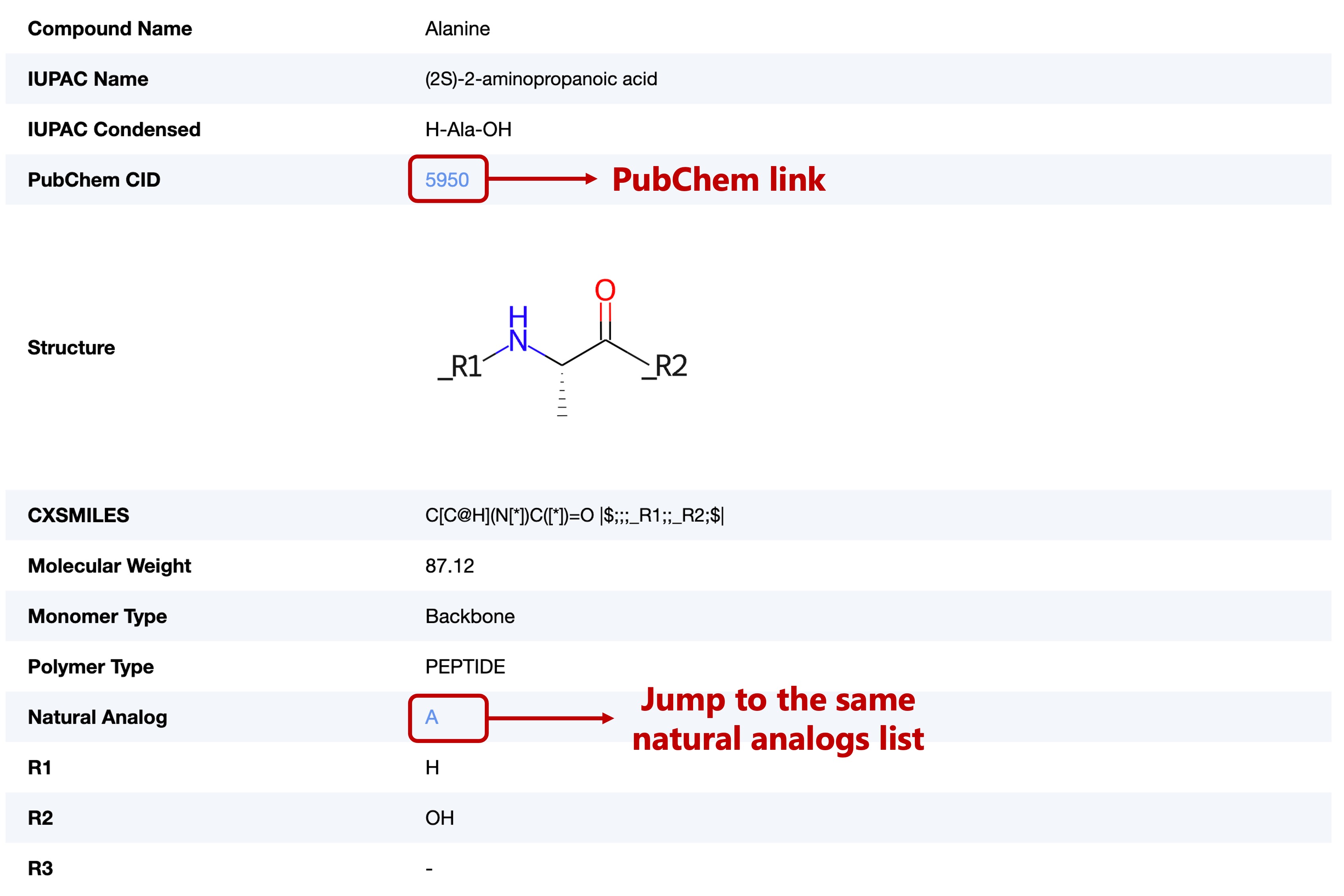 This section contains the basic information of each monomer, including:
This section contains the basic information of each monomer, including:

- Compound Name (from PubChem, source publication, etc.)
- IUPAC Name (from PubChem, and generated with STOUT software when not documented in PubChem)
- IUPAC Condensed (from PubChem)
- PubChem CID
- 2D Structure Image
- CXSMILES
- Molecular Weight (MolWt descriptor)
- Monomer Type
- Polymer Type
- Natural Analog (referenced the description of each monomer in PubChem and the monomer library of ChEMBL)
- Attachment Points (R1~R3)
3.3.2 Peptides Containing Current Monomer:
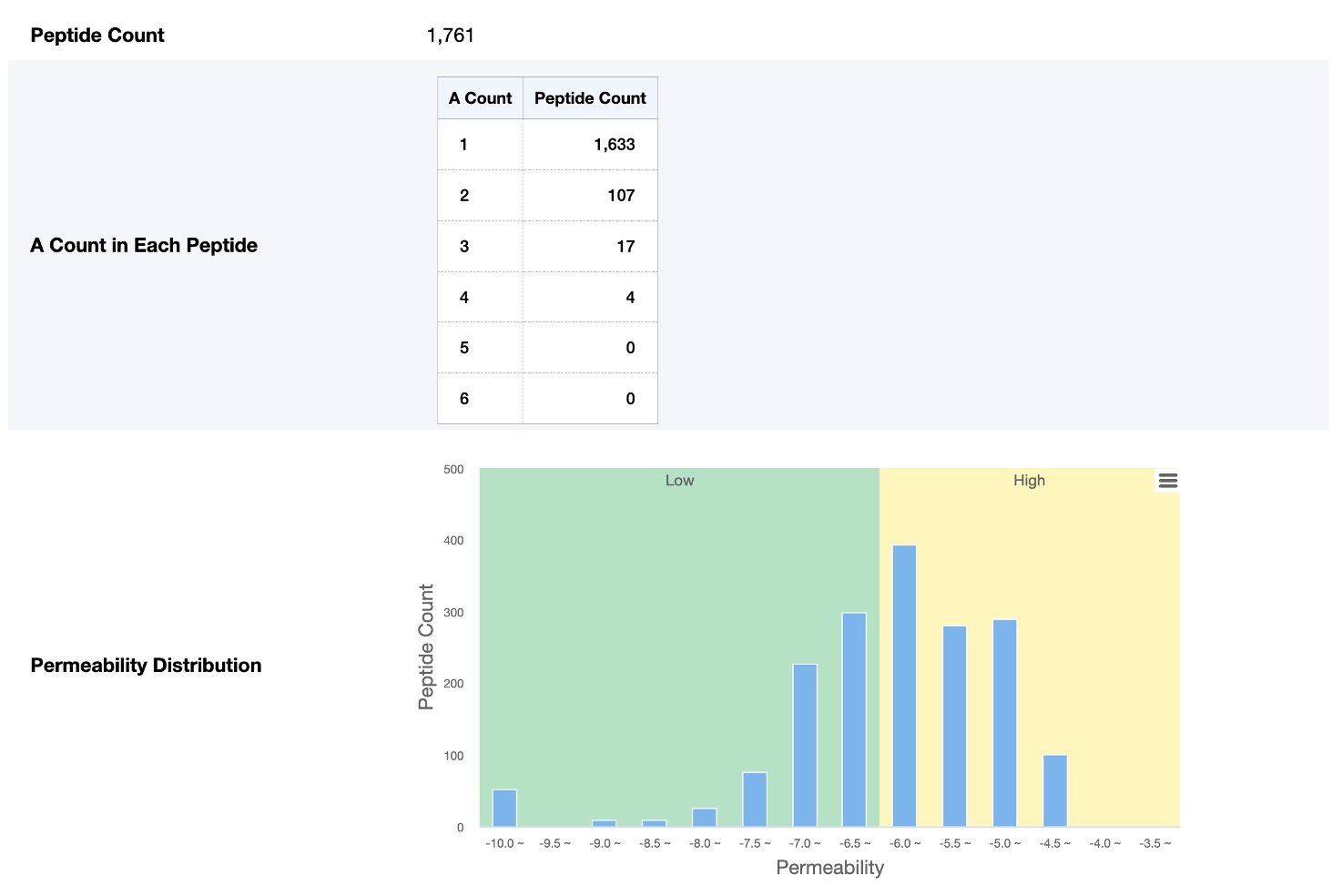 This section contains the peptides information which contains the current monomer, including:
This section contains the peptides information which contains the current monomer, including:

- Peptide Count (number of peptides containing at least one of the current monomer)
- Current Monomer Count in Each Peptide (number of peptides containing 1~6 current monomer respectively)
- Permeability Distribution
3.3.3 Physicochemical Properties:
This section contains six types of descriptor calculated by RDKit software for each monomer, including:
This section contains six types of descriptor calculated by RDKit software for each monomer, including:
- LogP (MolLogP descriptor)
- Ring Count (RingCount descriptor)
- Heavy Atom Count (HeavyAtomCount descriptor)
- Hydrogen Bond Acceptor Count (NumHAcceptors descriptor)
- Hydrogen Bond Donor Count (NumHDonors descriptor)
- Topological Polar Surface Area (TPSA descriptor)
4. Source Statistics
4.1 Literature List
CycPeptMPDB data were gathered from 54 published papers and 2
pharmaceutical company patents.
When collecting the data, we recorded not only basic information such as peptide structure
and membrane permeability, but also a detailed assay description.
You can see the information of these documents from Statistics part of the navigation bar.
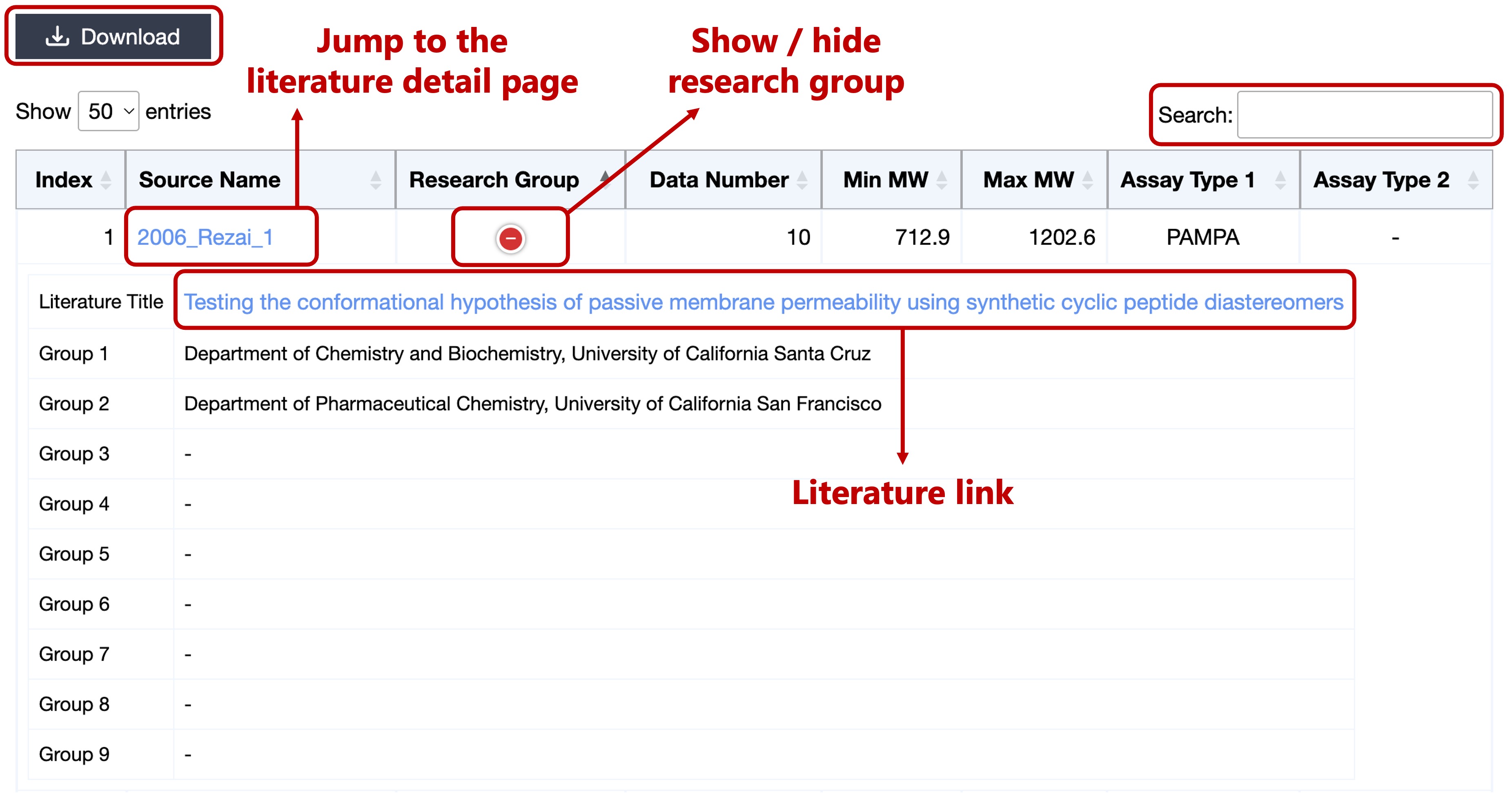
- Source Index
- Source Name
- Research Group (and Literature Title)
- Data Number
- Minimum Molecular Weight
- Maximum Molecular Weight
- Assay Types
Click on the Source Name can jump to the literature detail page.
Click on the Literature Title can jump to the original literature page.