
Peptide Search
Search Examples:
- Publication Year of Source: 2012 | <2016 | >2016 | 2012~2020
- Permeability: <-6 | >-6 | -6~-4
- Assay Type: PAMPA | Caco2 | MDCK | RRCK
- Original Name in Source: Cyclosporine | cyc | 1NMe3
- Molecular Weight: <800 | >800 | 800~1000
- Monomer Length: 6 | <6 | >6 | 6~12
- Molecule Shape: Circle | Lariat
- Combination: year:2008~2020, permeability:>-7, assay:PAMPA, weight:800~1200, length:>6, shape:Circle
About CycPeptMPDB
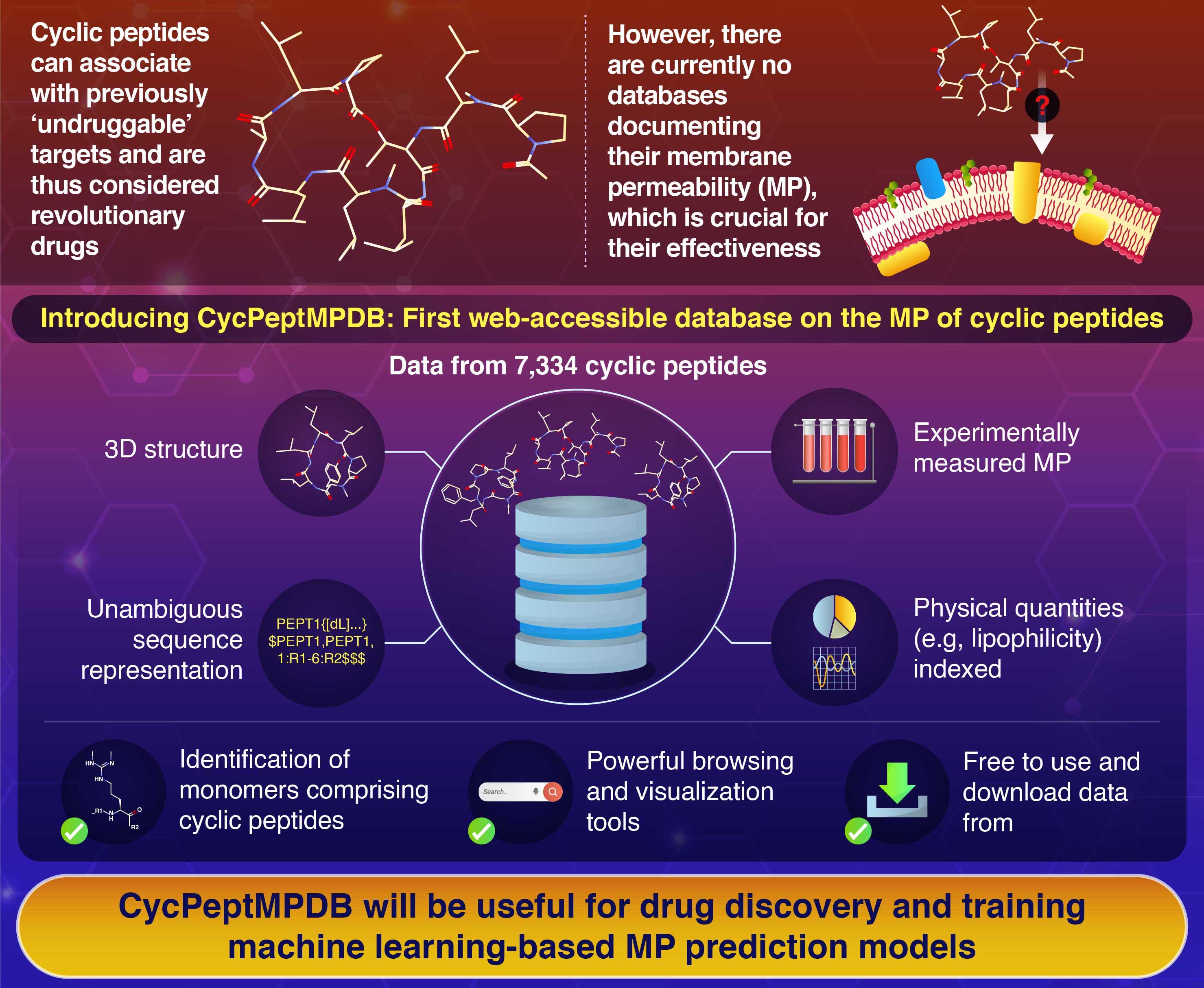
CycPeptMPDB
(Cyclic Peptide Membrane Permeability Database)
is the largest web-accessible
database of membrane permeability of cyclic peptide.
The latest
version 1.2
provides the information for
7,991
structurally diverse cyclic peptides
collected from
56
publications.
These cyclic peptides are composed of 385 types monomers (substructures).
As a comprehensive analysis platform,
CycPeptMPDB provides not only the basic information of
measured membrane permeability and structure,
but also provides
unified sequence representation (by HELM notation)
to unambiguously represent cyclic peptides.
In addition to data storage,
CycPeptMPDB provides several supporting functions such as online data visualization,
data analysis, and downloading.
CycPeptMPDB is expected to be a valuable platform to support membrane permeability research of cyclic peptides.
Update History
-
27-Dec-2024: Version 1.2
- 56 publications
- Added data from 2023_Ghosh, 2023_Ohta, 2023_Tanada, 2024_Bergeron, 2024_Faris, 2024_Kage, 2024_Koch, 2024_Ly, and 2024_Otani.
- 7,991 structurally unique cyclic peptides
- 8,466 entries (including duplicate structures); 8,880 permeability measurements
- 385 monomers
- Renamed monomer Mono7 as Bal(3-Me), Mono8 as Bal(d3-CF3).
- Redefined the natural analog of some monomers.
- Recalculated RDKit descriptors of peptides and monomers.
-
24-Apr-2023: Version 1.1
- Added 3D structure of cyclic peptides in chloroform selected based on the score given by a fast QM method.
- Added 3D structure of cyclic peptides in water selected based on the score given by a fast QM method.
- Added 3D-PSA values calculated from 3D structures in chloroform and water respectively.
- Special thanks to Dr. Richard A. Lewis of Novartis Institutes of BioMedical Research for providing the 3D structure and 3D-PSA calculation scripts.
- Methods for generating 3D structures will be available soon.
-
24-Feb-2023: Version 1.0
- 47 publications
- 7,334 structurally unique cyclic peptides
- 7,451 entries (including duplicate structures); 7,816 permeability measurements
- 312 monomers
- Reference: Li, J.; Yanagisawa, K.; Sugita, M.; Fujie, T.; Ohue, M.; and Akiyama, Y. CycPeptMPDB: A Comprehensive Database of Membrane Permeability of Cyclic Peptides, Journal of Chemical Information and Modeling, 63(7), 2240-2250, 2023. doi: 10.1021/acs.jcim.2c01573.